Estimation using abc rejection
Source:vignettes/estimation-using-abc-rejection.Rmd
estimation-using-abc-rejection.RmdSimple estimation using ABC-rejection
Create a reference trajectory
sum_stat_obs <- c(2.0, 0.75)Run abc smc procedure
res <- abcrejection(model_list = model_list,
prior_dist = prior_dist,
ss_obs = sum_stat_obs,
nb_acc_prtcl = 1000,
thresholds = 0.1,
max_attempts = 100000,
acceptance_rate_min = 0.01,
experiment_folderpath = "smplreject",
max_concurrent_jobs = 5,
verbose = TRUE)
#> Check folder_path for : smplreject/tmp
#> Folder created successfully.
#> Check folder_path for : smplreject/res
#> Folder created successfully.
#> Check folder_path for : smplreject/res/csv
#> Folder created successfully.
#> Check folder_path for : smplreject/res/figs
#> Folder created successfully.
#> Computation time - user : 5.802 s | system : 0.148 s | elapsed : 6.051 s
#> Experiment done!Plot results
all_accepted_particles <- res$acc_particles
all_tested_particles <- res$all_tested_particles
plot_abcrejection_res(all_accepted_particles, prior_dist,
filename="smplreject/res/figs/smplreject_pairplot.png",
colorpal = "YlGnBu")
#> [1] "Plot saved as '.png'."
plot_abcrejection_res(all_tested_particles, prior_dist,
thresholds=c(5.0, 4.0, 3.0, 2.0, 1.0, 0.1, 0.05, 0.01),
filename="smplreject/res/figs/smplreject_pairplot_custom_thresholds.png",
colorpal = "YlGnBu")
#> [1] "Plot saved as '.png'."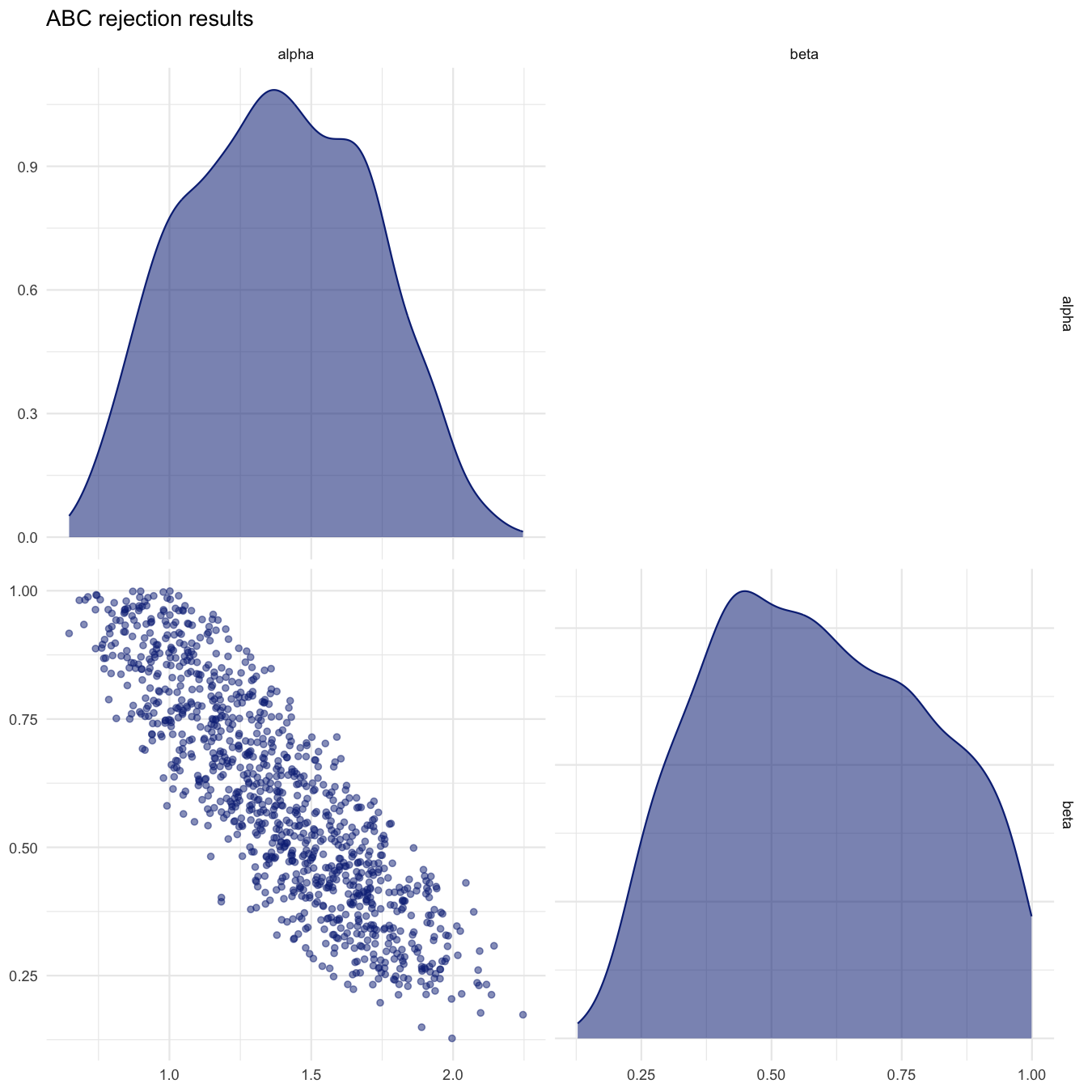
Pairplot of accepted particles for the predefined threshold
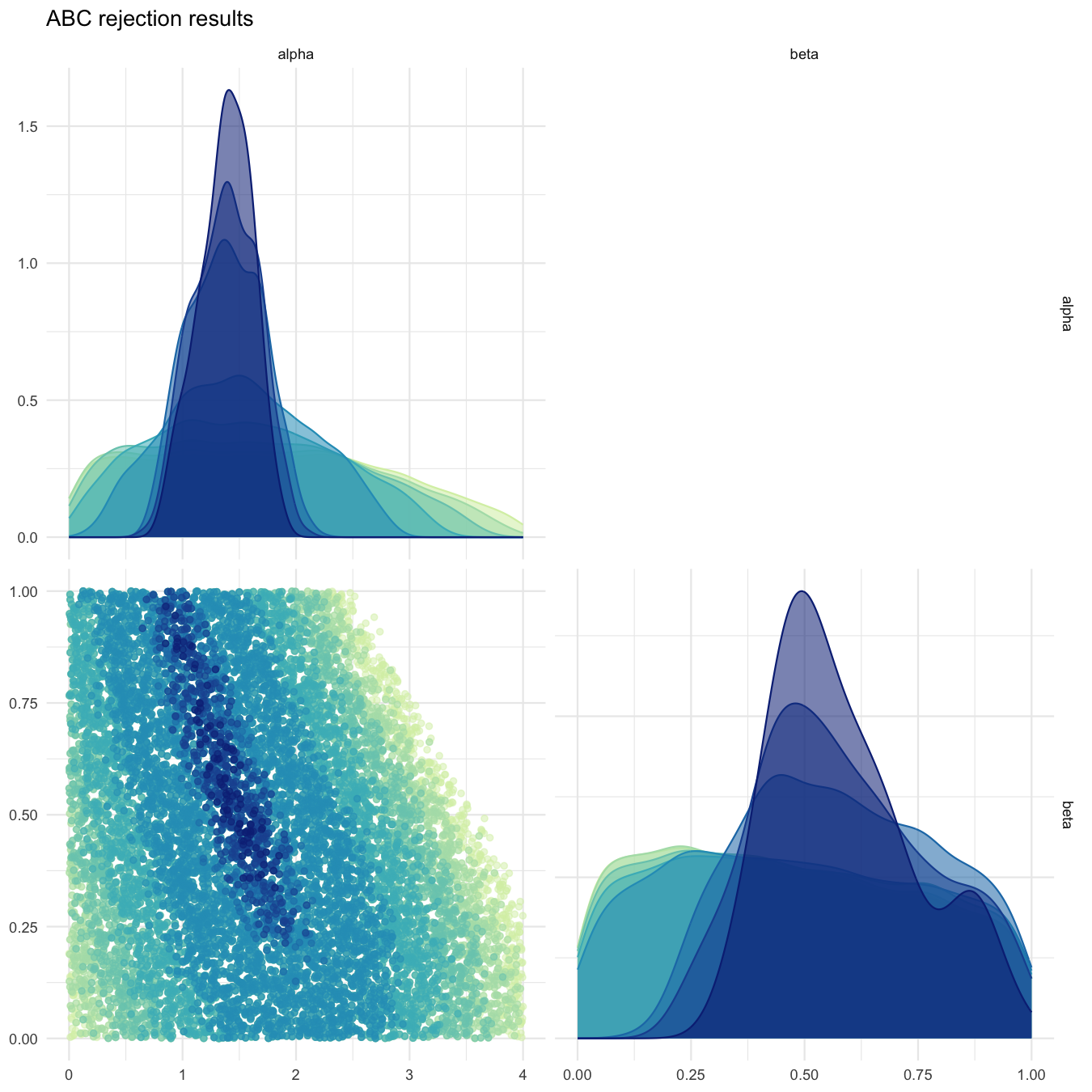
Pairplot of accepted particles for different acceptance thresholds