Using a model defined in a separate file
Source:vignettes/using-a-model-defined-in-a-separate-file.Rmd
using-a-model-defined-in-a-separate-file.RmdIn this example, the model is defined in a separate R file ‘model.R’:
toy_model <- function(x) {
c(x[["alpha"]] + x[["beta"]] + rnorm(1, 0, 0.1),
x[["alpha"]] * x[["beta"]] + rnorm(1, 0, 0.1))
}After that, the procedure is pretty much the same:
Create a reference trajectory
sum_stat_obs <- c(2.0, 0.75)Run abc smc procedure
res <- abcsmc(model_list = model_list,
model_def = "model.R",
prior_dist = prior_dist,
ss_obs = sum_stat_obs,
max_number_of_gen = 15,
nb_acc_prtcl_per_gen = 2000,
new_threshold_quantile = 0.8,
experiment_folderpath = "smpl",
max_concurrent_jobs = 5,
verbose = FALSE)Plot results
all_accepted_particles <- res$particles
all_thresholds <- res$thresholds
plot_abcsmc_res(data = all_accepted_particles, prior = prior_dist,
filename = "smpl/res/figs/smpl_pairplot_all.png", colorpal = "YlGnBu")
plot_densityridges(data = all_accepted_particles, prior = prior_dist,
filename = "smpl/res/figs/smpl_densityridges.png", colorpal = "YlGnBu")
plot_thresholds(data = all_thresholds, nb_threshold = 1,
filename = "smpl/res/figs/smpl_thresholds.png", colorpal = "YlGnBu")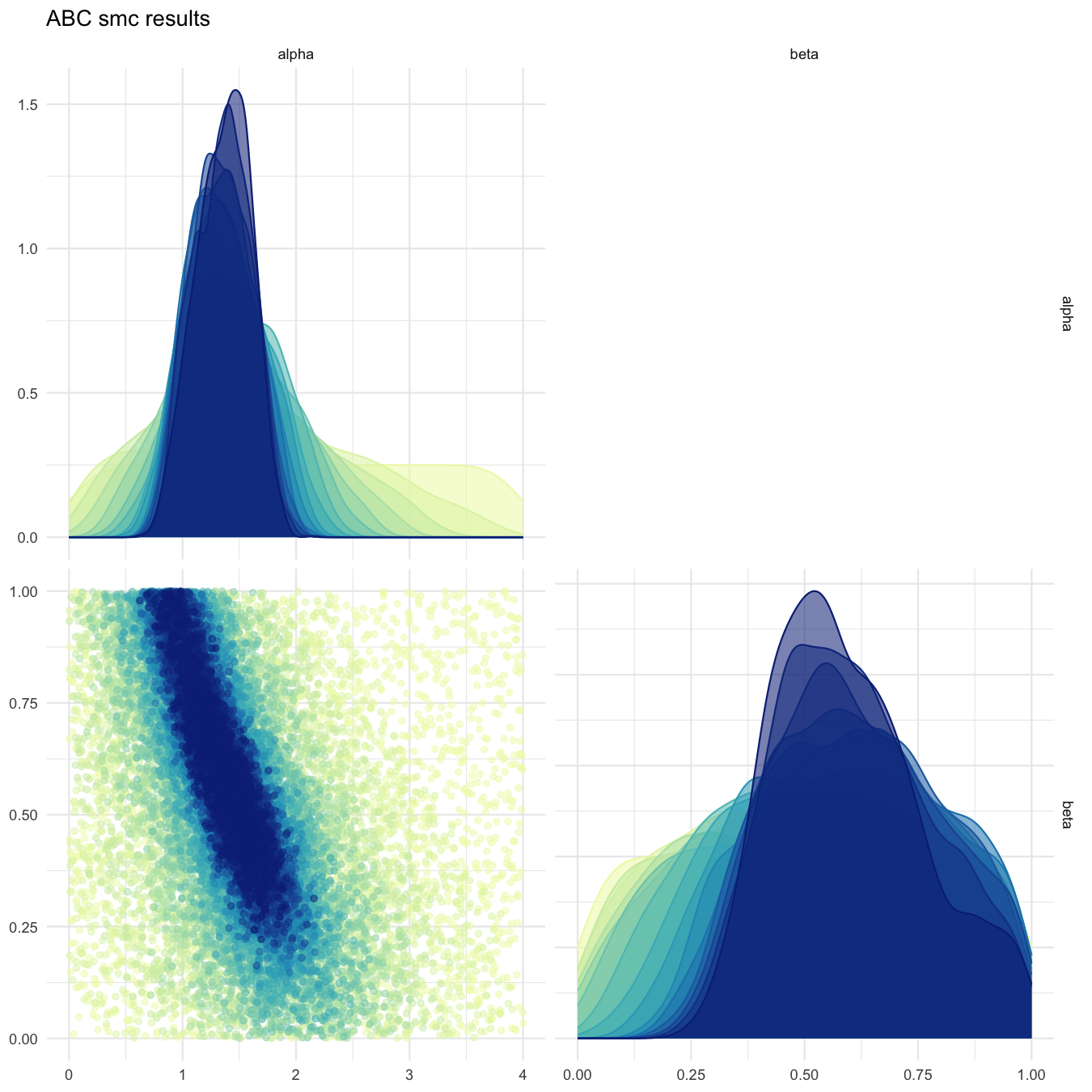
Pairplot of all iterations
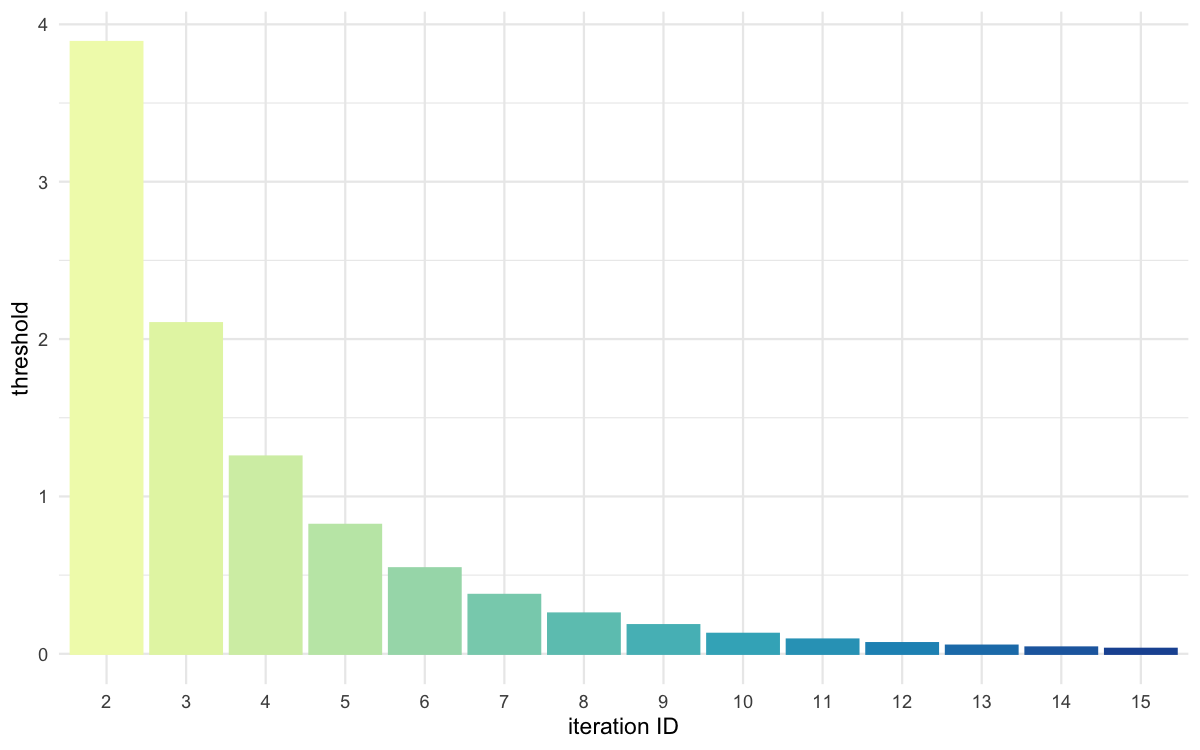
Threshold evolution over iterations
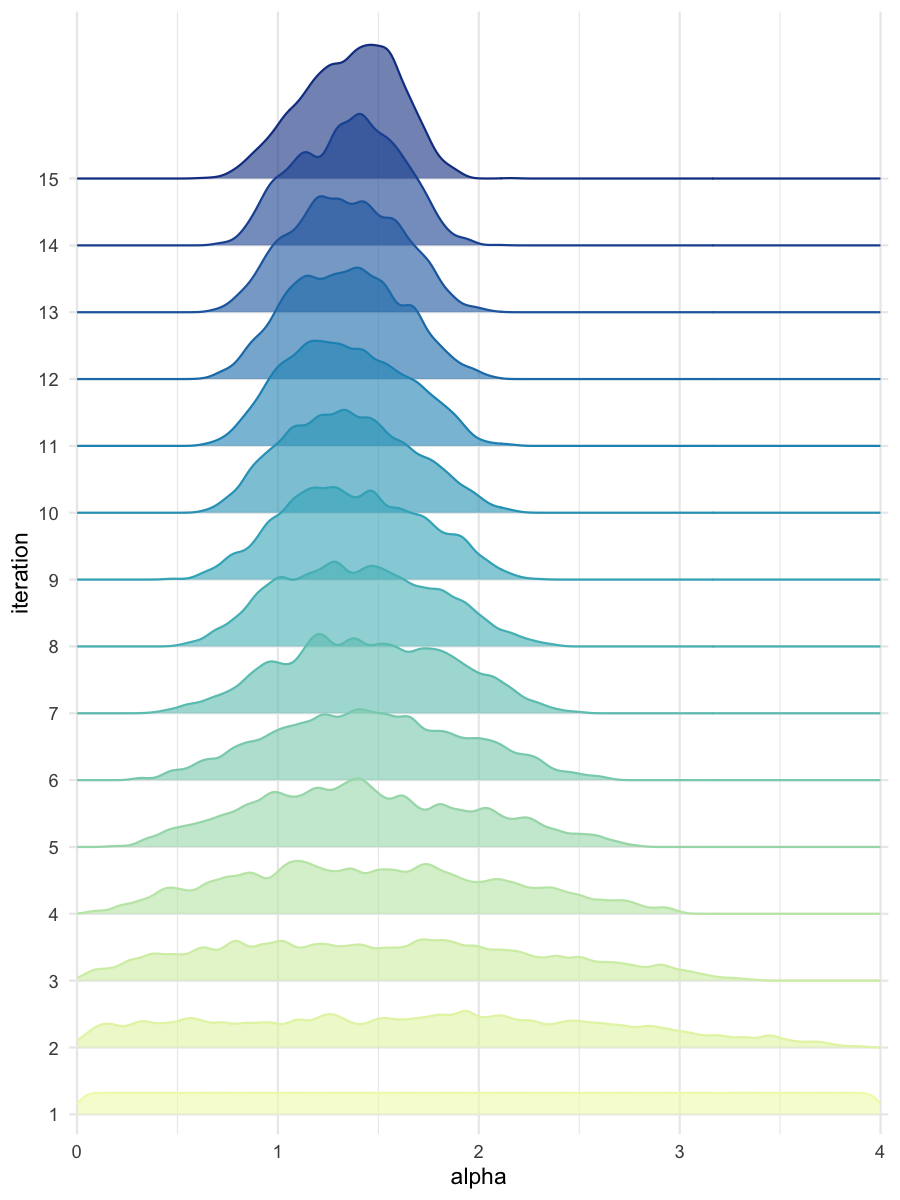
Density estimates for alpha
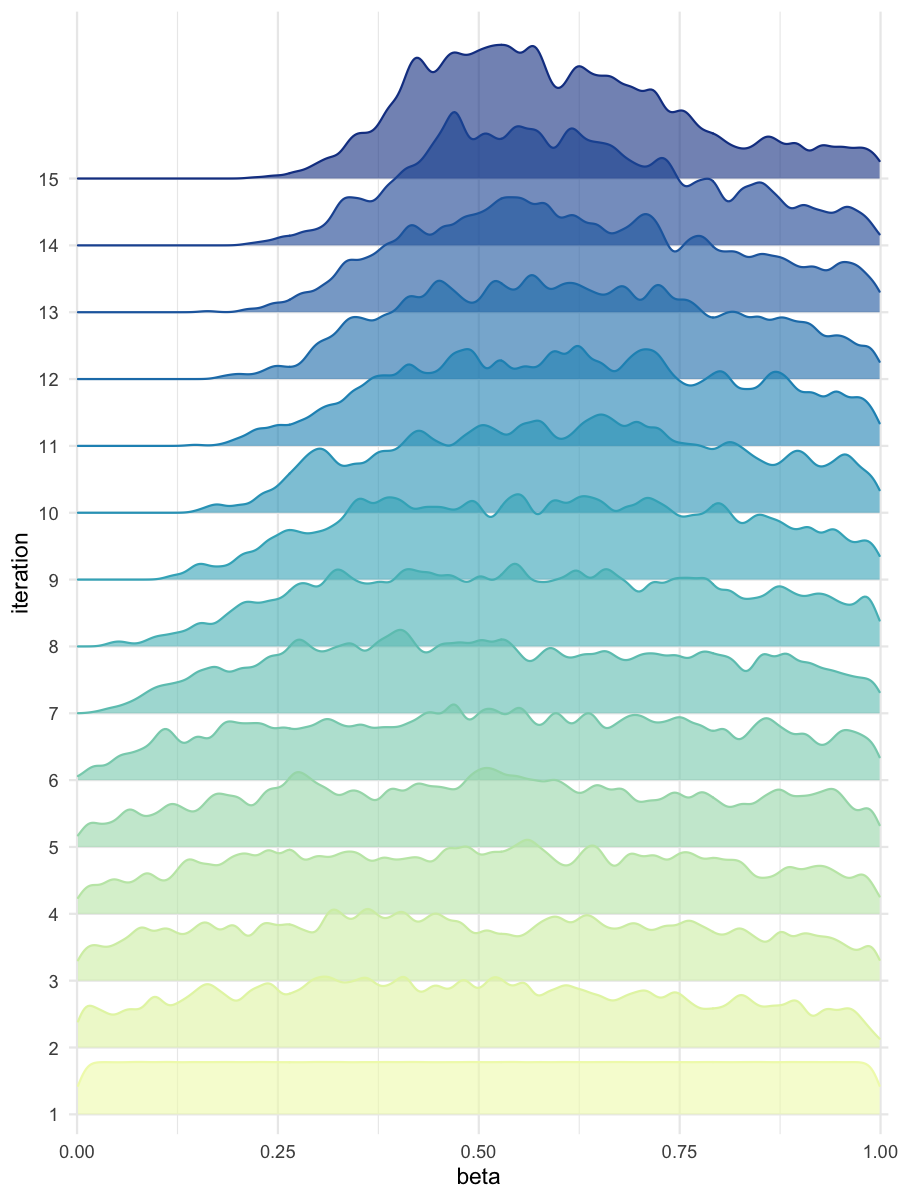
Density estimates for beta